
| Jialiang Huang, Ph.D. Professor E-mail: jhuang@xmu.edu.cn |
Education
2005, B.Sc., Fuzhou University
2008, M.Sc., Fuzhou University
2012, Ph.D., Chinese Academy of Science
Professional Experience
2011-2013, Scientist, GlaxoSmithKline (China) R&D Co., Ltd., Shanghai
2013-2018, Postdoc, Dana-Farber Cancer Institute/Boston Children's Hospital, Harvard Medical School
2018-present, Principal Investigator, School of Life Sciences, Xiamen University
Research Area
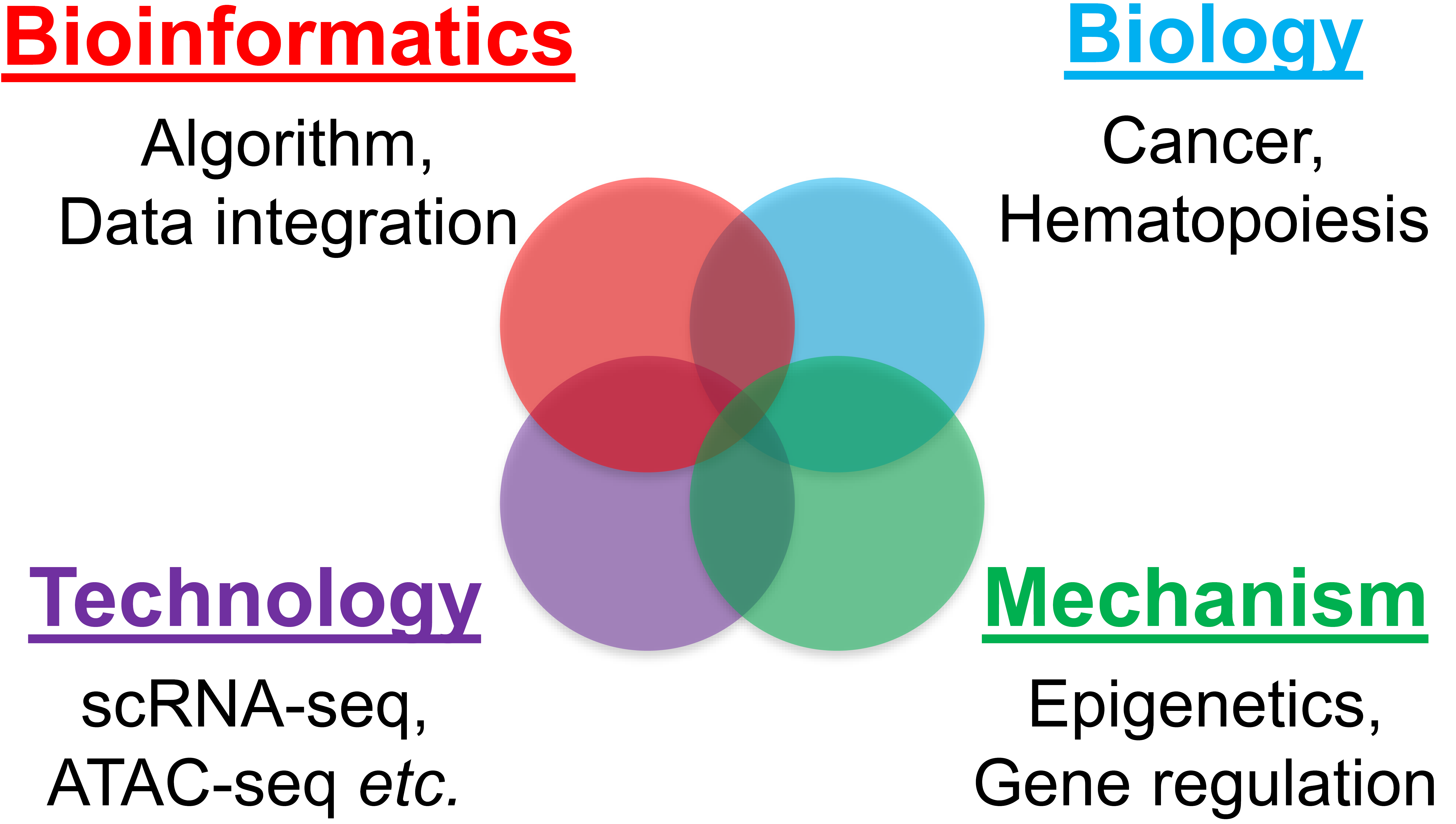
We are interest in understanding the epigenetic regulation mechanisms of development and cancer using computational (dry) and experimental (wet) methods. Our current research includes: (1) developing computational methods for quantification problems for various omics data, such as scRNA-seq, HiC, ATAC-seq and ChIP-seq; (2) integrating genomic, transcriptomic, and epigenomic data to elucidate the gene regulatory networks and for blood development; (3) using single-cell analysis and epigenetic approaches to investigate the cellular heterogeneity and gene regulatory mechanisms associated with cancer progression and treatment response, which helps to identify potential drug targets.
Selected Publications
1.M. Shu#, D. Hong#, H. Lin, J. Zhang, Z. Luo, Y. Du, Z. Sun, M. Yin, Y. Yin, L. Liu, S. Bao, Z. Liu, F. Lu*, J. Huang*, J. Dai*. Single-cell chromatin accessibility reveals enhancer networks driving gene expression during spinal cord development. Developmental Cell, 2022.
2.D. Hong#, H. Lin#, L. Liu, M. Shu, J. Dai, F. Lu, M. Tong, J. Huang*. Complexity of enhancer networks predicts cell identity and disease genes revealed by single-cell multi-omics analysis. Briefings in Bioinformatics, 2022.
3.Z. Wang#, T. Wang#, D. Hong#, B. Dong#, Y. Wang, H. Huang, W. Zhang, B. Lian, B. Ji, H. Shi, M. Qu, X. Gao, D. Li, C. Collins, G. Wei, C. Xu*, H. J. Lee*, J. Huang*, J. Li*. Single-cell transcriptional regulation and genetic evolution of neuroendocrine prostate cancer. iScience, 2022.
4.Y. Kai#, B. E. Li#, M. Zhu#, G. Y. Li, F. Chen, Y. Han, H. J. Cha, S. H. Orkin, W. Cai*, J. Huang*, G.-C. Yuan*. Mapping the evolving landscape of super-enhancers during cell differentiation. Genome Biology, 2021.
5.W. Cai#, J. Huang#, Q. Zhu, B. Li, D. Seruggia, P. Zhou, M. Nguyen, Y. Fujiwara, H. Xie, Z. Yang, D. Hong, P. Ren, J. Xu, W. Pu, G.-C. Yuan* and S. H. Orkin*. Enhancer-dependence of cell-type-specific gene expression increases with developmental age. PNAS, 2020.
6.J. Huang#, K. Li#, W. Cai, X. Liu, Y. Zhang, S. H. Orkin, J. Xu*, G.-C. Yuan*. Dissecting super-enhancer hierarchy based on chromatin interactions. Nature Communications, 2018.
7.E. Marco#, W. Meuleman#, J. Huang#, K. Glass, L. Pinello, J. Wang, M. Kellis*, G.-C. Yuan*. Multi-scale chromatin state annotation using a hierarchical hidden Markov model. Nature Communications, 2017.
8.J. Huang#, X. Liu#, D. Li#, Z. Shao#, H. Cao, Y. Zhang, E. Trompouki, T. V. Bowman, L. I. Zon, G.-C. Yuan, S. H. Orkin*, J. Xu*. Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Developmental Cell, 2016.
9.J. Huang, E. Marco, L. Pinello, G.-C. Yuan*. Predicting chromatin organization using histone marks. Genome Biology, 2015.
10.J. Huang, C. Niu, C. D.Green, L. Yang, H. Mei*, J.-D. J. Han*. Systematic prediction of pharmacodynamic drug-drug interactions through protein-protein-interaction network. PLoS Computational Biology, 2013.
11.J. Huang, Y. Liu, W. Zhang, H. Yu and J.-D. J. Han*. eResponseNet: a package prioritizing candidate disease genes through cellular pathways. Bioinformatics, 2011.